Overview of the server
The interface of DisCanVis offers multiple convenient approaches for users to find relevant data. The homepage contains a general description of the web server with direct links to the Getting started, Examples and API pages, where more detailed information is available. There are four main tabs that provide access to the search, browse, help and statistics pages.
The Search page allows users to query the database using different terms derived from the UniProt and COSMIC databases: Accession, entry, full and partial names from the UniProt database and identifiers from the COSMIC database are all accepted. When multiple entries are found, another search box appears to further refine search terms.
Tables group data based on certain properties and provide direct links to entries together with their names and identifiers and some basic statistics about the mutations collected with respect to either a selected region or the whole protein, depending on the table. Each column can be filtered based on matching worlds. These fields accept basic regular expression based searches as well. For example, in the Significantly Mutated table, we can exclude mucin proteins by putting “!muc” in the Uniprot name box (without quotes). Numerical values can be restricted by > and < signs. For example, to select proteins more than 100 residues long, place “>100” in the sequence length box.
More about search options: tablesorter
The first table labeled “Drivers” contains known cancer drivers. “Experimental disorder” table contains proteins with experimentally verified disordered regions derived from the MobiDB database with either “curated” or “homology” level evidence. The third option called “Binding domain” contains proteins with linear peptide binding domains collected from the PixelDB, DIBS and ELM databases. The “ELM” and “ELM Switches” tables contain proteins with regions obtained from the Eukaryotic Linear Motif Database Instances and Switches section respectively. The last two tables enable users to select entries based on chromosome or “GO term”, respectively.
Statistics page contains some results of the overall analysis of the functional and structural annotation of the mutations from our database. The first graphs are about the disordered/ordered distribution from different features. The last plots show genes with the most mutated known ELM motifs and known ELM switches motifs.
Features
The main goal of our visualization tool is to integrate mutational data with genome and protein level information, as both levels are important to interpret the functional and structural impact of the observed genetic variations. The following features are included in the visualization and can also be accessed through REST API.
- sequence - amino acid sequence of the protein
- exon - exon boundaries
- phastcons - genome conservation values ranging from 0 to 1, according to the PhasCons method
- complexity-seg - low complexity regions according to the SEG method
- complexity-dust- low complexity regions according to the DUST method
- complexity-trf - low complexity regions according to the TRF method calculated from the CDS sequence
- polymorphism - all and common polymomphisms collected from the dbSNP database collected and classified by UCSC Genome Browser
- omim - disease mutations collected from the Humsavar dataset
- clinvar - disease mutations collected from the ClinVar database
- pdb - known protein structure corresponding to given entry
- pfam - pfam sequence families for a given protein. Motifs are colored by gray. Domains are colored by green. Families are colored by lightblue. Repeats are colored by orange. Coiled-coils are colored by dark blue. Disordered regions are colored by pink.
- anchor - disordered binding regions predicted according to ANCHOR method using a cutoff value 0.5 (orange - disordered binding regions, light blue - not a disordered binding region)
- iupred - disorder prediction according to IUPred (version, parameters!)
- mobidb - experimentally verified disordered regions collected through the MobiDB databases
- alphafold - alphafold pLDDT scores
- tcgam - missense mutations from the TCGA projects
- tcgaf - truncating mutations (frameshift and nonsense mutations) from the TCGA projects
- tcgai - short in-frame insertions and deletions from the TCGA projects
- cosmicm - missense mutations from COSMIC
- cosmicf - truncating mutations (frameshift and nonsense mutations) from COSMIC
- cosmici - short in-frame insertions and deletions from COSMIC
- roisig - Significantly mutated regions calculated with ISimpre on TCGA mutations.
- elm - short linear motifs from the ELM database
- elmswitches - molecular switches from the switches.ELM database
- ptm - post-translational modifications from Uniprot and PhosphositePlus databases
- roi - Region of Interest from UniProt database.
- binding - Binding regions from UniProt database.
- dibs - Disordered binding regions from DIBS database.
- mfib - Mutual Folding Induced by Binding from MFIB database.
- binding_domain - known binding domains that recognize disordered regions
- phasepro - regions involved in driving phase separation
- conservation - position-specific conservation values at the main evolutionary levels
Visual features
The main goal of our visualization tool is to integrate mutational data with genome and protein level information, as both levels are important to interpret the functional and structural impact of the observed genetic variations.
Each entry has a header section. The header shows the protein name according to UniProt. By clicking on the icon left of the protein name, additional details are shown about the entry, such as the gene name, the chromosome, and length of the protein sequence. In addition, the UniProt accession and ENSEMBL transcript ID are also shown with links to the corresponding databases. On the right hand side of the header section, the cancer driver status is given.
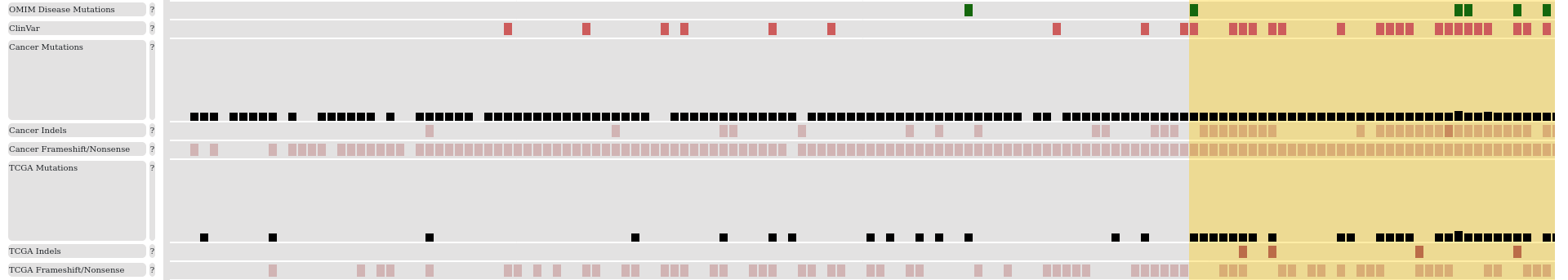
By clicking on the icon next to this information, the source of this categorization can be accessed. The next few icons enable the user to select features that are shown in more detail, to select cancer types in the mutation profile and to present a brief summary table about the mutations. The final icon returns to the homepage of the database. The top of the page also contains an overview of the whole protein which shows the cancer mutations, domains and the combined disorder information along the sequence. There is a slider that can be moved along the sequence, indicating the region shown in more details below.
The detailed information for the selected region can be divided into four main sections. The first section presents the position indicators and the sequence and additional information that can be helpful to assess the relevance of variations. These include for example exon boundaries which can be useful to identify mutations located at splicing sites. We also show repeat regions identified by the TRF method. Such regions often show increased mutational rates. In our previous work, we found that low genomic conservation calculated by the PhastCons method is a good indicator of regions that contain more mutations without likely disease relevance. We also indicate polymorphisms that commonly occur in the human population. In general, mutations that occur in repeat regions, have low genomic conservation or coincide with common polymorphisms are likely to correspond to passenger mutations.
The next section presents genetic variations: both general disease mutations and specific cancer mutations. We collected pathogenic “Disease” variants from The UniProt Humsavar database (version 2022.02.) with links to the OMIM database. We also incorporated ClinVar variants labeled as Pathogenic/Likely Pathogenic. Currently, cancer mutations generated by the TCGA projects are shown. Single amino acid change variations are shown as bars, with the height of the bar proportional to the number of mutations in the given position. In-frame indels and truncating (frameshift and nonsense) mutations are shown separately. In these cases color intensity is proportional to the number of observed variations. On the top of the page, cancer types can be restricted and this filtering is also reflected in the header section. Mutations collected from the COSMIC database, which includes both large-scale and targeted studies, can also be accessed, but are not shown by default. We also highlight significantly mutated regions. Driver mutations often accumulate in specific regions, especially when a large number of samples are analyzed together, while passenger mutations are expected to be distributed evenly along the sequence. To identify such regions, we used the iSiMPRe method. The main advantage of iSiMPred is that it can automatically find boundaries of regions that are enriched in mutations, without prior definition of region of interest.
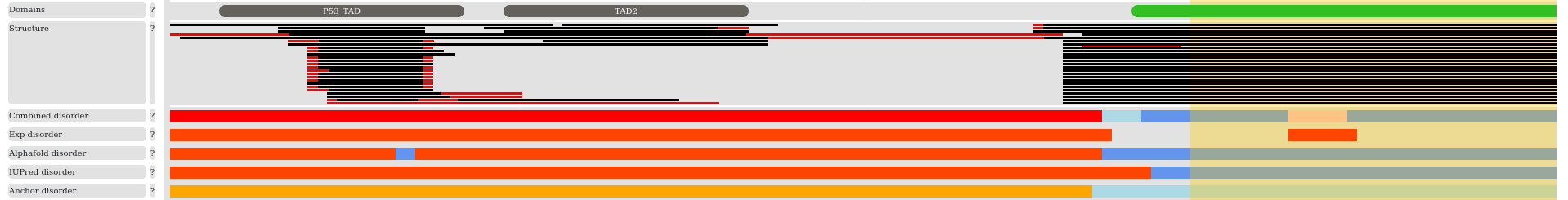
The third section presents various information about the structural state of proteins. We show known domains according to the PFAM database (2022.08). The structures collected from the PDB. The red lines in the structures indicate missing residues in case of X-ray structures and mobile regions in case of NMR structures. In most cases such regions indicate protein disorder. Information about experimentally verified disordered regions are transferred from the MobiDB database. We also included prediction methods, such as IUPred , the pLDDT scores of the AlphaFold2 method and the ANCHOR prediction to highlight disordered binding regions. In general, disordered regions are indicated by red, while ordered regions with blue color. Even experimental annotations can be wrong and predictions obtained with different methods often contradict each other. We developed a combined disorder approach to help to reconcile these cases. This method is based on a simple decision tree and favors experimental methods, highly confident pLDDT predictions, and IUPred predictions in this order.
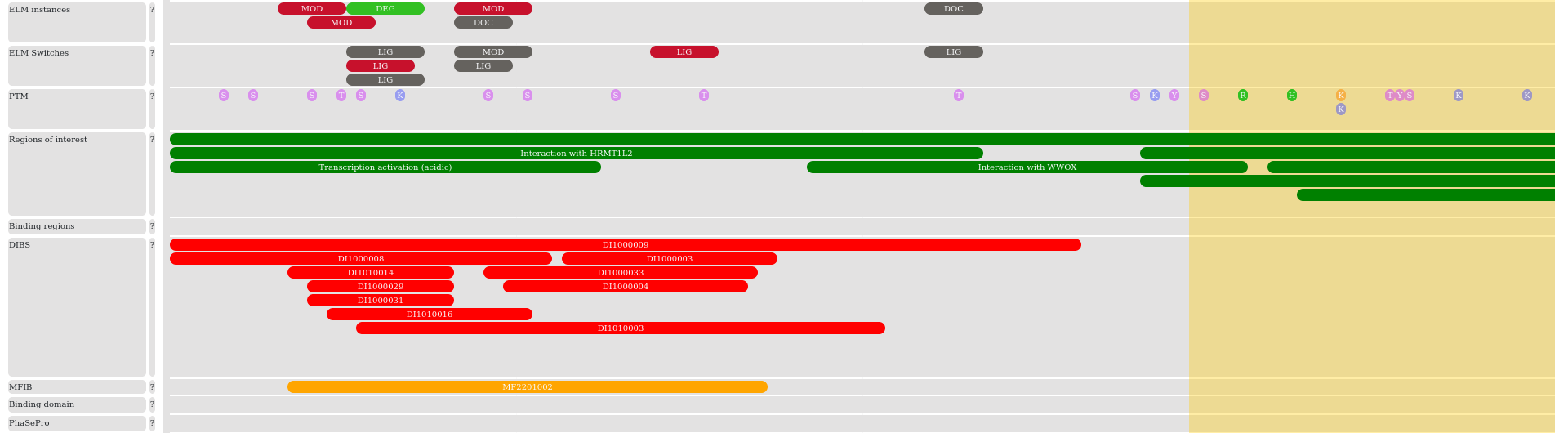
We collect regions of interest and binding region annotations directly from the SwissProt database. We added short linear motifs and SLiM switches from the ELM databases. Disordered regions that undergo coupled folding and binding by interacting with globular proteins or another disordered protein are collected from the DIBS and MFIB databases, respectively. All these regions can be selected and are linked to their source databases. Post-translational modifications from dbPTM and PhosphositePlus are also indicated, using different representations for phosphorylation, acetylation, methylation and ubiquitination. In addition, we also show regions that are involved in driving the formation of membraneless organelles through phase separation based on annotations in the PhaSePro database.
The final section presents position specific sequence conservation calculated from multiple sequence alignment of orthologs. To enable the mapping of the conservation scores to the query protein sequence, deletion-free alignments were used.
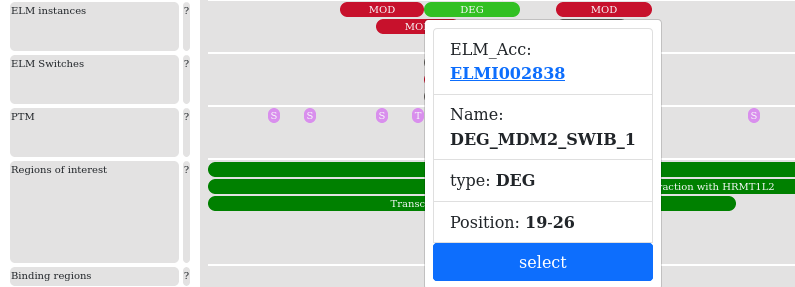
The visualization tool has interactive features to help focus on specific regions and its annotations. The functional boxes are either clickable and or hoverable. Clickable functions show a popover on click, where some detailed information can be found. All clickable boxes can be selected with a vertical box by either on click or clicking the “select” button in the popover. Clicking on the same button will deselect the region. Most of the functional and structural annotation popover boxes contain links to the corresponding database with the specific id.
Updates
Version 1.2 - 2023.05.11
Added + Modified: Mutational data
- COSMIC mutation
- TCGA mutation from COSMIC database
- Added TCGA mutation from TCGA Legacy database
- Added CBioportal mutation
Added: PPI data
- Intact database
- Biogrid database
- Hippie database
Added: Browse tables
- OMIM Diseases
- Phase Separation
How to cite
DisCanVis: Integrating structural and functional annotations for the better understanding of the effect of cancer mutations located within disordered proteins.
Norbert Deutsch, Mátyás Pajkos, Gábor Erdős, Zsuzsanna Dosztányi Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest, Hungary
zsuzsanna.dosztanyi@ttk.elte.hu
Published: Protein Science journal, 30 November 2022 doi: https://doi.org/10.1002/pro.4522